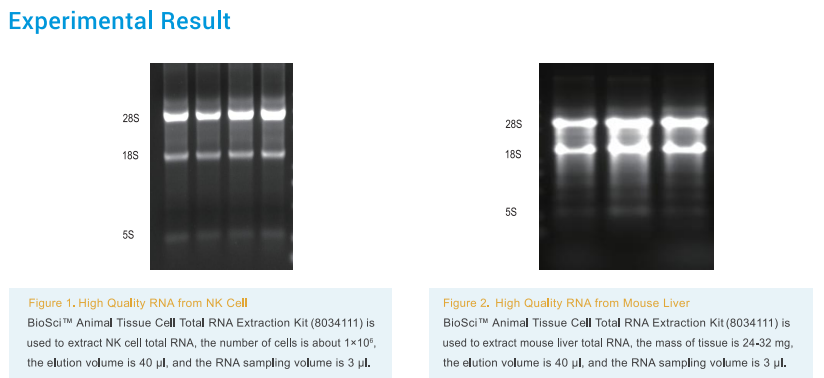
Introduction
Isolating total RNA from animal tissues is a basic step in many molecular biology techniques, including studying gene expression, RNA sequencing (RNA-seq), and reverse transcription quantitative PCR (RT-qPCR). Getting high-quality RNA is essential for these techniques to produce reliable data. There are several methods to extract total RNA from animal tissue cells, each with pros and cons.
Common RNA Extraction Methods
Organic Solvents:
This traditional method uses phenol-chloroform extraction. It works well but involves hazardous chemicals and requires experience to get good results.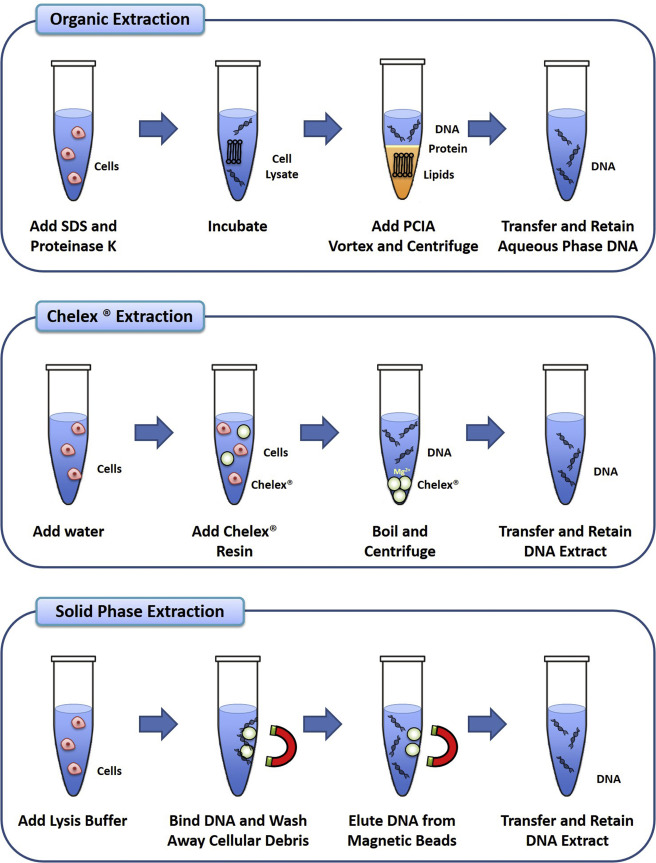
Tri Reagent:
This commercially available solution combines the properties of phenol and chloroform, making extraction simpler. However, Trizol can be expensive and may not work for all tissues.
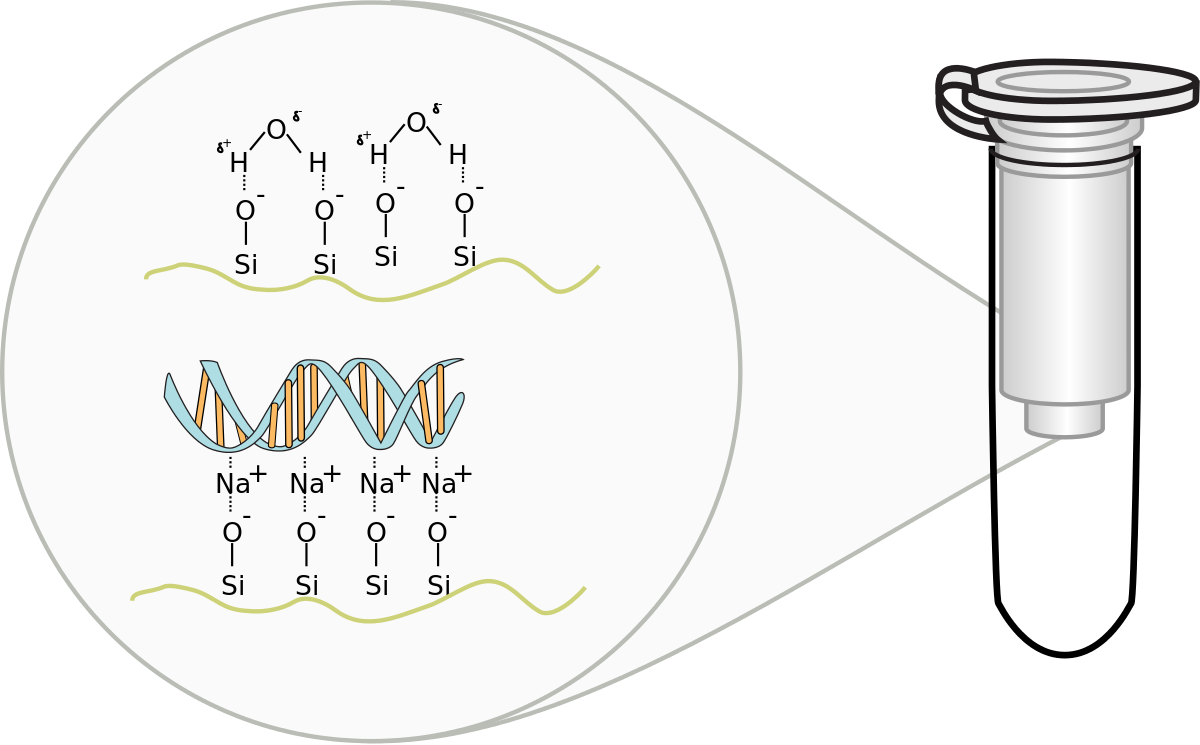
Silica-based Columns:
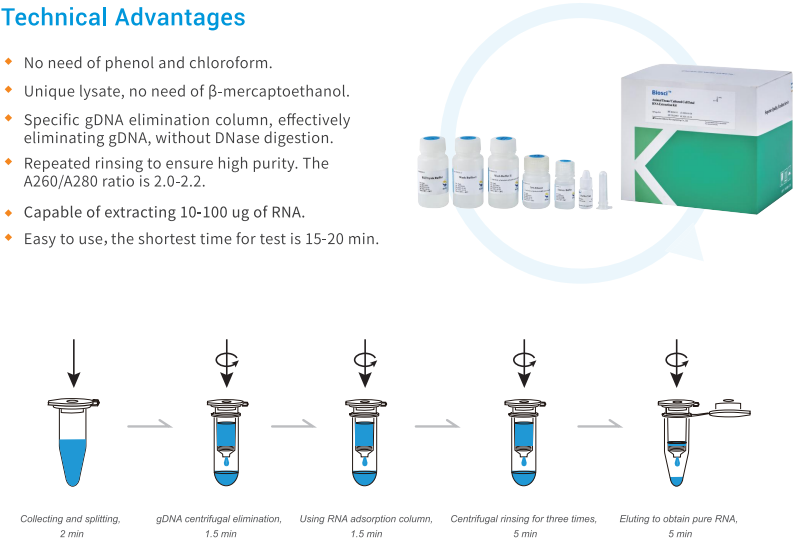
This popular method uses silica-membrane spin columns to purify RNA. It offers high purity, good yields, and works with various tissues. However, some silica column kits require adding carrier RNA during extraction to recover small RNAs efficiently.
Factors Affecting RNA Extraction
Several factors can influence the success of RNA extraction from animal tissues. The type of tissue itself plays a role, as RNA content and composition can vary between tissues. Some tissues may require protocol optimization to account for challenges like the presence of fibrous components or enzymes that degrade RNA (RNases). Proper handling of tissue samples before and during RNA extraction is critical. Techniques like snap-freezing or using RNA stabilization solutions can help minimize RNA degradation. Finally, the downstream application can also influence method selection. For instance, RNA sequencing requires highly purified RNA with minimal degradation, whereas RT-qPCR may tolerate RNA with slightly lower purity. Researchers must consider these factors to ensure they choose the optimal RNA extraction method for their specific experiment.
Several factors can influence the success of RNA extraction from animal tissues. The type of tissue itself plays a role, as RNA content and composition can vary between tissues. Animal Tissue Cell Total RNA Extraction Kits address this challenge by providing optimized protocols and reagents specifically formulated to handle the unique properties of different tissue types. These kits often include homogenization buffers to efficiently lyse cells and release RNA, while also minimizing the activity of RNases that degrade RNA. By following the optimized protocols and using the specialized reagents provided in Animal Tissue Cell Total RNA Extraction Kits, researchers can ensure efficient extraction of high-quality RNA from a variety of animal tissues. This ensures reliable results in downstream applications like RNA-seq or RT-qPCR.
Conclusion
The best method for animal tissue cell total RNA extraction depends on various factors such as the tissue type, the amount and purity of RNA needed, and budget constraints. Researchers should consider these factors and consult relevant scientific literature when choosing an RNA extraction method for their specific research project.